Abstract
Clonal hematopoiesis (CH) is a ubiquitous phenomenon of human aging - nearly all individuals older than 50 years have CH in a small but detectable fraction of their blood. Currently, the mechanisms driving clone evolution are poorly understood and experimental models recapitulating the emergent behavior of spontaneously-arising low-frequency clones (early CH) are lacking. To address this gap in knowledge, we endeavored to establish the mouse as a model for native clonal hematopoiesis.
Laboratory mice live only 2-3 years but CH in humans develops over decades; thus, we considered whether natural CH would be observable outside of manipulated transplantation models. To address this question, we performed computational modeling incorporating published estimates of murine hematopoietic stem cell (HSC) mutation rate and population size. This analysis suggested that clonal expansions are likely to occur in mice, although at magnitudes-smaller clone sizes than in humans. Thus, we hypothesized that natural murine CH would be characterized by rare and small somatic clones.
To test murine blood for CH, we employed targeted duplex consensus sequencing. In this strategy, each initial DNA molecule is uniquely barcoded such that reads derived from complementary starting strands are resolved to error-corrected sequences, enabling magnitudes-greater sensitivity relative to most other sequencing methods. We developed a panel to probe for rare variants in mouse homologs of human CH driver genes and queried unmanipulated, naturally-aged, laboratory mice across their lifespan, at ages spanning 4 to 37 months (roughly equivalent to humans between 20 to 100 years of age).
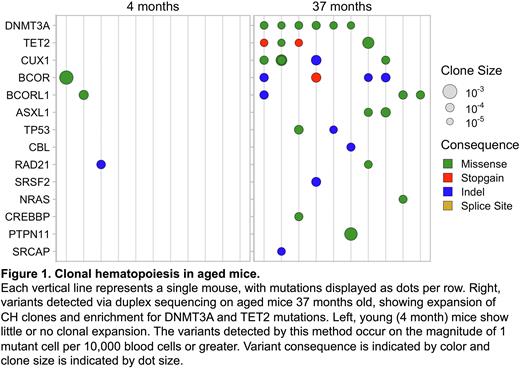
Remarkably, we readily observed expanded variant clones in aged mice, with rare or absent clonal expansions in young mice (Figure 1). CH incidence increased with each year of age. Clones were most commonly driven by somatic mutations in known CH-associated genes, such as Dnmt3a, Tet2, and Bcor/Bcorl1. These variants occurred on the order of 1 mutant cell per 10,000 blood cells, often aligned with human hotspot loci, and were enriched for non-synonymous mutations above what is expected by genetic drift. Longitudinal sampling demonstrated the persistence of clones during aging and following transplantation. Thus, natural murine CH is broadly similar to that observed in humans, with the exception of the size of clones, as aligned with our predictions.
Most laboratory mice are maintained throughout their life in highly controlled conditions with very low pathogen exposure, in stark contrast to the constant varied microbial exposures of humans. Because inflammation has been suggested to accelerate CH, we considered the possibility that a more natural microbial environment would augment murine CH.
To test this, we employed a "normal microbial experience” (NME) model, in which laboratory mice are infected with common mouse microbes via exposure to fomite bedding (sourced from pet store mice), subjecting young and aged mice to short-term NME. Strikingly, we observe an increased number and size of expanded clones in NME-exposed aged mice relative to specific pathogen-free housed controls. Importantly, clones bearing mutations in Trp53 were particularly enriched in NME-exposed mice, as measured by clone count and clone magnitude (allele fraction). NME exposure transmits multiple types of pathogens; thus it is challenging to identify specific effects. Therefore, we performed targeted exposure to Mycobacterium avium, which we have previously shown can activate HSCs. We again observed an exacerbation of clonal expansion in aged, infected mice, with specific enrichment for Bcor/Bcorl1 mutant clones.
In summary, we have unequivocally demonstrated that aged mice develop CH naturally within their captive lifespan. Genes and even mutational hotspots found in human CH are recapitulated, and the magnitude and number of clones are aligned with predictions based on mutation rate and HSC number. Moreover, native murine CH is significantly accelerated in the context of microbial exposure, revealing that driver genes offer context-specific advantages towards expansion. Our approach detects low-frequency clonal interplay and will be applicable for the development of therapeutic strategies to modulate CH by shifting the hematopoietic fitness landscape as a whole, rather than by targeting specific CH clones.
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal